Let it shine
The light-based technique optogenetics has revealed marvels in neuroscience. In the past 10 years, researchers have used the technique to, among other things, make lab animals , and elicit the , all on command.
The method has been hailed as a revolution in neuroscience. In 2010, the journal Nature Methods declared optogenetics to be . In 2013, the prestigious Grete Lundbeck European Brain Research Prize went to six researchers whose work is considered to have laid the foundations for the technique (see box). Optogenetics even made its way into the Obama administration’s and the .
Now some scientists want to see if optogenetics can repeat its magic in understanding how cells beyond the ones in the brain work. “The hope is that optogenetics is going to enable transformations in cell biology as it’s already enabled in neuroscience,” says at the University of California, San Francisco.
Biologists are starting to test out optogenetics to understand the nuances of basic cellular processes, such as migration, polarity, transcription and signal transduction. “We’ve seen an explosion of interest in the idea of controlling biological activity with light,” says of Stanford University, whose laboratory is creating light-controlled proteins to study nucleotide exchange and protease activities.
As recognized by the Brain Prize, several groups contributed to the development of optogenetics. But most people mention a 2005 Nature Neuroscience paper as the one that launched a thousand optogenetics experiments. (There are more than 1,100 papers in PubMed with the word “optogenetics.”)
The Nature Neuroscience paper, authored by , now at Massachusetts Institute of Technology, and at Stanford University and others, described the introduction of a gene for an algal ion channel called channelrhodopsin-2 into rat hippocampal neurons. The light-sensitive ion channels opened up when researchers shone light on the neurons and triggered synaptic transmission. The investigators effectively were in control of when the neurons fired.
The work made neuroscientists sit up: This was a way to selectively control neurons of their own choosing, which they couldn’t do with other approaches. After watching experiment after experiment come out in neuroscience based on optogenetics, at Princeton University says, cell and molecular biologists “looked at all that success and reasoned, if you can activate specific neurons in a large population, maybe… you can also activate a specific signaling protein in a large complicated network that acts within a cell.”
 Human cells grown in six-well plate for an optogenetics experiment in an incubator fitted with an LED panelLAURA B. MOTTA-MENA, ELIZABETH ORTH AND KEVIN H. GARDNER, CUNY ADVANCED SCIENCE RESEARCH CENTER
Human cells grown in six-well plate for an optogenetics experiment in an incubator fitted with an LED panelLAURA B. MOTTA-MENA, ELIZABETH ORTH AND KEVIN H. GARDNER, CUNY ADVANCED SCIENCE RESEARCH CENTER
Like needle-nose pliers
It has been tough to understand the contributions of individual components in complicated molecular networks. While many experimental perturbations of cells have yielded important information about biology, in most cases their mode of action is much like that of a hammer smashing the entire cell.
Electrodes for electrical stimuli poke into cells; the mere poking can trigger a response even before the experiment begins. Some chemical stimuli can bind indiscriminately to untargeted molecules and often are irreversible. Gene knockouts permanently prevent production of related proteins, sometimes causing dramatic effects that aren’t physiologically relevant. Knockout experiments also are slow, allowing cells time to compensate for the missing gene before scientists can characterize the effects of the knockout.
Light is different. Researchers can place a light source a distance away from the experimental system, eliminating the effects of touch. Most importantly, with lasers and LEDs, researchers can control the amount of light and the duration of light exposure, which lends greater precision to protocol measurements. Light exposure also is reversible, as researchers can switch back and forth between light and dark.
When researchers combine light, with all these useful attributes, with the expression of specific genes in cells, they get a technique with the exactness of a pair of fine-tipped, needle-nose pliers: Researchers make a group of cells artificially express photosensitive proteins. When these cells are exposed to light, photosensitive proteins activate, allowing researchers to see directly what effects these proteins have on a set of cells or even a whole animal.
The technique “is such a different way of approaching science than just knocking proteins out or overexpressing them with a (green fluorescent protein) tag on them,” says Weiner. It “enables us to plug in the intermediate steps of a cascade so we can isolate a given module with spatial and temporal control.”
The ability to reach in and play with a single molecule among a mishmash of hundreds of molecules is one of optogenetics’ most appealing attributes. Because light beams can be directed to specific locations in the cell, optogenetics gives scientists the ability to investigate the influence of a protein’s coordinates. Experts interviewed for this story cited the ability to study cell dynamics as another major appeal.
Information on location and timing of cellular events makes optogenetics different from other light-based analytical methods. In optogenetics, light acts “more as the actuator than as the sensor,” driving the pace and place of the experiments, says at the University of Colorado. “You really can’t do that through any other approach. That’s where I feel optogenetics has a lot of power.” Other light-based approaches rely on the light for sensing, in that fluorescent reporter molecules passively emit photons, reporting cell behavior.
Experts interviewed for the story point to experiments that have been hard to do or impossible by other means but that now seem doable with optogenetics. Toettcher gives the example of p53, a well-known tumor suppressor that’s turned on in response to several stimuli. But researchers realized that the protein doesn’t turn on and stay on in response to a particular stimulus. For something like ionizing radiation, p53 fluctuates in pulses over time. In fact, the number of pulses of p53 is proportional to the amount of damage the cell receives. Unfortunately, most molecular biology techniques gloss over the time-dependent expressions of proteins. Western blots simply detect the average expression of proteins, and gene knockout experiments annihilate all expression or artificially jack up expression.
“There is a much more sophisticated code at work in cell biology than people originally thought,” says Toettcher. “That’s really where these optogenetic tools in cell signaling are going to make a big impact. They allow us, for the first time, to dissect the contributions of different dynamic responses.”
The Brain Prize
The , also known as the Brain Prize, recognizes original and influential advances in any area of neuroscience. In 2013, the prize went to Gero Miesenböck, Ernst Bamberg, Peter Hegemann, Georg Nagel, Ed Boyden and Karl Deisseroth. The award announcement cited these six men for laying the foundations of optogenetics.
The prize recognized Bamberg, Hegemann, Miesenböck and Nagel for making the fundamental observations and discoveries about light-sensitive molecules and developing them so they could be introduced into specific types of neurons. It acknowledged Boyden and Diesseroth for developing the technique further and getting it to work in live animals. Colin Blakemore, chairman of the foundation’s selection committee, said in a press release: “Optogenetic control of nerve cells is arguably the most important technical advance in neuroscience in the past 40 years. It offers a revolution in our understanding of the way in which circuits of neurons carry out complex functions, such as learning and controlling movement. And it could provide an entirely new approach to the restoration of function in blindness or brain degeneration, and to the treatment of a variety of other neurological and psychiatric disorders.”
LOV and more
At the core of optogenetics are the genes for light-sensitive proteins that researchers plop into cells. The field heavily relies on photosensitive molecules, such as channelrhodopsins and other opsins, derived from microorganisms and plants. One set of molecules has come from the light-oxygen-voltage-sensing domains, better known as LOV domains, which are found in plants, algae, fungi and bacteria.
at The City University of New York Advanced Science Research Center and colleagues have engineered a bacterial LOV-containing protein called EL222. When illuminated with blue light, EL222 binds to DNA in less than 10 seconds in several mammalian cell lines and intact zebrafish embryos.
This timescale of seconds is important. “When you knock proteins down or out, (the experiment) takes on the order of days, weeks or months to perform. Cells have ways of upregulating compensatory pathways,” notes Weiner. “But if you can take them out on a timescale of seconds, you can see phenotypes that are invisible with the slower perturbations.”
LOV domains also have been used to study the influence of place. at the University of Utrecht and his team could be used to attach cytoskeletal motor proteins to peroxisomes, mitochondria and recycling endosomes to move them around to different parts of cells. For example, Kapitein and colleagues demonstrated that the local positioning of recycling endosomes influenced the growth of neurons.
Another optogenetics approach is to turn a protein that’s oblivious to light into a version that responds to light. at the University of North Carolina and his group did just that in 2009 with . “Instead of harnessing the functions of naturally occurring molecules, molecules can now be manipulated to do things that nature never engineered them to do,” says Hahn.
The GTPase Rac regulates the actin cytoskeleton. The photoactivatable version of Rac changes conformation when hit with light and becomes active. Researchers now can see in detail how photoactivated Rac influences different parts of the actin cytoskeleton. Because Rac controls cell movement, researchers have been able to use photoactivatable Rac to guide living cells to precise positions inside living animals and to start to work out how cell placement influences phenomena such as development and the trafficking of immune cells in and out of tumors.
With these engineered light-sensitive proteins, explorations inside cells become more sophisticated. Weiner gives the example of the Ras/MAP kinase cascade, which regulates a range of biological processes, such as cell differentiation, proliferation and arrest. Researchers have been puzzled by how one cascade can regulate such different biological outcomes. But with optogenetics, researchers can drive the activation of the cascade directly and begin to figure out how sustained and transient activations and dosages regulate cell biology.
In 2013, Weiner, along with Toettcher, who was his postdoctoral fellow at the time, and UCSF’s Wendell Lim, used the plant Phy-PIF system to delve deeper into the . Phy, a phytocytochrome, photoswitches between two different states; PIF binds to one of those two states. The investigators attached Phy to the membrane and fused PIF to various activators, such as a protein called SOS, of the Ras pathway in the cytoplasm. When they hit Phy with a red light, it was able to interact with PIF; when they hit Phy with infrared light, the interaction went away.
By playing around with different doses and timed pulses of light, the investigators determined, for example, that the transcription factor STAT3 is activated by only a particular part of the cascade after photoactivatable Ras is active for more than an hour.
Removing the kinks
For all their enthusiasm for optogenetics, experts recognize the glitches. “The initial hope with optogenetics was it was going to be as easy as GFP. You just slap it on the end of your favorite molecule and regulate its activity,” says Weiner. “It turns out it’s harder than that.”
One reason optogenetics has been less plug-and-play in molecular biology than it has been in neuroscience is that every neuron is “activated by the exact same kind of stimulus, which is membrane depolarization,” says Toettcher, but, in other cells “it’s not at all the case that every signaling protein is activated by the same kind of input.”
Another problem dogging the field is inconsistency: One LOV domain may work like gangbusters in a particular cell line or on a particular protein but drop dead in another. “I hate to say it, but there’s a ton of variability right now,” says Gardner. “Those of us who are developing these tools and are the first adopters really need to start thinking about addressing this a little more comprehensively.”
Gardner applauds colleagues who have published papers, which he describes as “not sexy,” that systematically work through different photosensitive proteins to see what exactly they can do. Some of the proteins “work great everywhere. Some work great in some cell lines in some settings and not others,” says Gardner. “Some have been tough to adopt by people not born out of the discoverer’s lab.”
Then comes the question of physiological relevance. As with any method that is based on changing the cell, how do researchers know that they are not rendering the system useless by the simple act of introducing a foreign light-controlled protein? “You have to make sure you’re not altering the physiology of the cells before you turn on the light,” says Hahn. “That’s challenging.”
There is also the issue known as “leakiness.” That’s when a light-sensitive protein is residually active without light, causing changes in the cell even before the experiment begins. Hahn says the issue can be addressed by measuring the level of activity of the protein as it gets different doses of light so a researcher knows how much activity is present in the absence of light or, better yet, can build a protein that has a large difference in activity when lit and when not lit.
Once researchers are armed with better photosensitive modules, the aim will be to integrate several of them into a single experiment. The multiplexing aspect “in theory, sounds great because there are already proteins out there with different colors,” says Hahn. But it hasn’t been possible so far because “none of them are really perfect.” Hahn and others state that improving the proteins so that they turn on and off cleanly with a specific wavelength of light is valuable, but grunt, work.
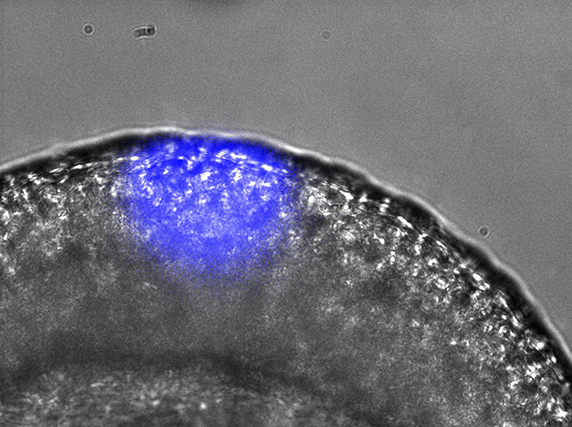 Gene expression can be spatially controlled by optogenetics in the zebrafish embryo.ANNA READE, STEPHANIE WOO AND ORION WEINER, UCSF.
Gene expression can be spatially controlled by optogenetics in the zebrafish embryo.ANNA READE, STEPHANIE WOO AND ORION WEINER, UCSF.
Gonna leave the light on
Investigators using optogenetics in molecular biology say the kinks will be ironed out as more people try out optogenetics in their experimental systems. They say no special skills are needed, although engineering new optogenetics approaches does require specialization in protein engineering and optics. Some savviness with cloning and protein expression, a bit of microscopy savoir faire, and you’re able to harness the powers of optogenetics. There isn’t even a need for highly specialized microscopes or complicated lasers.
Experts are willing to lend a helping hand. Weiner puts out the welcome mat: “I hope people interested in using optogenetics will fly out to our lab for a few days or a week to learn how it works. It’s much easier to learn and take back the technique from our lab than reading protocols and trying to troubleshoot over the phone.”
Adopters of optogenetics feel that the effort invested into the technique will pay off simply because it allows nuanced, delicate measurements within complicated systems. Besides the ability to tease out the effects of place and timing, researchers can analyze the effects of doses of protein activity by exploiting the amount of light fed into an experimental system. “That’s one thing the field isn’t taking much advantage of yet is the fact that it’s titratable,” says Weiner. “You can not only acutely control signals in time, but you can dose them in a titratable, reversible fashion for more quantitative biology.”
So experts feel their enthusiasm for optogenetics is warranted. “The number of applications is just screaming for attention. A lot of people are getting very creative here,” says Gardner. There is no doubt in his mind that the next five years “will be an interesting period.”
Enjoy reading ASBMB Today?
Become a member to receive the print edition four times a year and the digital edition weekly.
Learn moreGet the latest from ASBMB Today
Enter your email address, and we’ll send you a weekly email with recent articles, interviews and more.
Latest in Science
Science highlights or most popular articles

Elucidating how chemotherapy induces neurotoxicity
Andre Nussenzweig will receive the Bert and Natalie Vallee Award at the 2025 ASBMB Annual Meeting, April 12–15 in Chicago.

Where do we search for the fundamental stuff of life?
Recent books by Thomas Cech and Sara Imari Walker offer two perspectives on where to look for the basic properties that define living things.

UCLA researchers engineer experimental drug for preventing heart failure after heart attacks
This new single-dose therapy blocks a protein that increases inflammation and shows promise in enhancing muscle repair in preclinical models.

The decision to eat may come down to these three neurons
The circuit that connects a hunger-signaling hormone to the jaw to stimulate chewing movements is surprisingly simple, Rockefeller University researchers have found.

Curiosity turned a dietitian into a lipid scientist
Judy Storch will receive the Avanti Award in Lipids at the 2025 ASBMB Annual Meeting, April 12–15 in Chicago.

From receptor research to cancer drug development: The impact of RTKs
Joseph Schlessinger will receive the ASBMB Herbert Tabor Research Award at the 2025 ASBMB Annual meeting, April 12–15 in Chicago.

