JBC: Tor comes to the fore in autophagy
Healthy cells are expert recyclers, rapidly breaking down worn-out or surplus macromolecules and reusing their building blocks. Several pathways, such as the proteasomal degradation route for protein breakdown, specifically pick out those damaged or expendable molecules.
 Yoshinori Ohsumi and colleagues discovered that the Tor protein controls in yeast cells. Government of Japan/Wikimedia Commons
Yoshinori Ohsumi and colleagues discovered that the Tor protein controls in yeast cells. Government of Japan/Wikimedia Commons
These selective degradation pathways usually operate in cells that have ready access to nutrients. But when cells encounter crises such as severe nutrient shortages, they activate an emergency recycling strategy. In these cells, a bulk degradation pathway indiscriminately breaks down macromolecules and entire cytoplasmic organelles and ribosomes.
This process is called autophagy (Greek for “self-eating”) — or sometimes, more specifically, macroautophagy to distinguish it from specialized forms of autophagy that are more restricted and selective. It’s the rough equivalent of a wood chipper that shreds chairs, tables and credenzas wholesale for use in plywood or as fireplace fuel.
Autophagy also represents that eliminates long-lived proteins and damaged organelles in some cells, such as neurons, whose total destruction by other processes such as apoptosis would harm the organism.
Defects in autophagy have been linked to neurodegenerative diseases and cancer, highlighting that this recycling mechanism is essential for keeping cells healthy and alive. However, how the cellular nutrient status is communicated to the autophagy machinery was long unknown.
In published in the Journal of Biological Chemistry and now recognized as a JBC Classic, Takeshi Noda and Yoshinori Ohsumi at the National Institute for Basic Biology in Japan uncovered this missing link, reporting that the target of rapamycin, or Tor, protein in yeast suppresses autophagy in cells that are growing in nutrient-rich conditions.
A facile biological system
Researchers had known since the early 1990s that two Tor proteins of Saccharomyces cerevisiae, or budding yeast, respond to the nutritional state of the cell and regulate cell cycle progression. Noda and Ohsumi’s discovery that Tor also controls autophagy was a milestone in unraveling how bulk degradation is regulated in eukaryotic cells.
Ohsumi in part because it has a large vacuole (a structure analogous to the lysosome in mammalian cells) that is easy to study by light microscopy. In 1992, that in nutrient-starved cells of yeast mutants that cannot degrade proteins via the proteasomal pathway, the vacuoles quickly fill up with spherical structures.
These membrane-bound structures contained ribosomes, mitochondria, lipids and cytosolic enzymes, suggesting that they were signs of autophagy; the researchers named them autophagic bodies.
Noda, the first author of the Classic paper, was a graduate student in the Ohsumi lab. During peer review of the 1992 paper, he said, the referees asked for additional experiments to verify that what he and Ohsumi saw under the microscope was indeed autophagy.
The extra work paid off. “We clearly showed that cytoplasmic enzymes are incorporated into the isolated vacuole during this process,” Noda said; these additional findings convinced the referees and secured the paper’s publication.
The referees’ requests prompted Noda to develop more specific and sensitive methods. “After the experience with the first paper, I realized that we needed a quantitative assay for measuring autophagy activity,” he said.
A revised view
In , Noda and others in the Ohsumi lab developed an assay that measures the activity of a genetically engineered alkaline phosphatase, or ALP, that becomes active only when moved to the yeast vacuole as it is during autophagy.
“Having this system in hand, I got interested in studying the regulatory mechanism of autophagy in more detail,” Noda said.
Noda came across from Michael Hall’s group at the University of Basel, Switzerland, reporting that Tor in yeast controlled cellular processes that typically are induced in response to starvation.
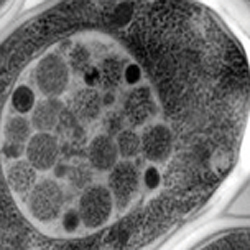 When yeast cells are depleted of nutrients, autophagic bodies (small dark spherical structures) start to accumulate in their vacuoles (light gray structure). Noda and Ohsumi showed that the Tor protein inhibits autophagy in nutrient-rich conditions.Journal of Biological Chemistry“As autophagy is induced by starvation, I was eager to use our ALP assay to test the hypothesis that Tor might also regulate autophagy,” Noda said.
When yeast cells are depleted of nutrients, autophagic bodies (small dark spherical structures) start to accumulate in their vacuoles (light gray structure). Noda and Ohsumi showed that the Tor protein inhibits autophagy in nutrient-rich conditions.Journal of Biological Chemistry“As autophagy is induced by starvation, I was eager to use our ALP assay to test the hypothesis that Tor might also regulate autophagy,” Noda said.
Noda and Ohsumi grew yeast cells in nutrient-rich culture conditions that typically inhibit autophagy. The researchers then added the Tor inhibitor rapamycin to the cell cultures. Using their ALP-based autophagy assay, they detected autophagy in the Tor-inhibited cells, and using light microscopy, they also observed the tell-tale autophagic bodies in the vacuoles.
Using a yeast strain that carried a mutated TOR2 gene encoding a temperature-sensitive Tor2 variant, the authors firmed up these observations, finding that the TOR2-mutant strain exhibits autophagy when grown at temperatures that inactivated the mutated Tor2 variant.
“Our discovery of Tor’s role in autophagy revised prevailing views at the time, showing that Tor regulates not only anabolic processes, but also catabolic ones,” Noda said. “This coupling of anabolic and catabolic processes has been a key game changer in the research field.”
In search of a mechanism
They next turned to the question of how Tor might control autophagy. The Ohsumi lab 14 autophagy-deficient yeast strains, and Noda found that rapamycin did not restore autophagy in these mutants, indicating that these genes, called ATG, all act downstream of Tor.
The Classics paper could not resolve the question of how Tor might regulate the ATGs, but there was a promising hint — to have a kinase domain essential for cell cycle regulation. This suggested that Tor might control autophagy by phosphorylating and thereby inhibiting autophagy-associated proteins such as the ATGs.
This was borne out by published by Ohsumi and colleagues reporting that Tor phosphorylates the autophagy-related yeast protein Atg13, interfering with Atg13’s ability to activate another kinase, Atg1, required for autophagy induction.
reviewed how using the facile yeast system has helped pinpoint many other key players in autophagy.
Despite great strides in clarifying autophagy mechanisms in both yeast and mammalian cells, more work is needed. In particular, to prevent self-destruction, cells must avoid excessive degradation during autophagy, Noda said, meaning they need to have mechanisms that terminate autophagy at the right time.
“We are currently investigating this question,” Noda said, “and already got some good candidate molecules” that might stop autophagy. For work that helped uncover the molecular mechanisms in autophagy, Ohsumi was awarded the Nobel Prize in physiology or medicine in 2016.
JBC Associate Editor Paul Fraser at the University of Toronto nominated the paper as a Classic. Read this and other JBC Classics on the journal's .
Enjoy reading ASBMB Today?
Become a member to receive the print edition four times a year and the digital edition weekly.
Learn moreGet the latest from ASBMB Today
Enter your email address, and we鈥檒l send you a weekly email with recent articles, interviews and more.
Latest in Science
Science highlights or most popular articles

From the journals: MCP
Protein analysis of dopaminergic neurons. Predicting immunotherapy responses in lung cancer. ZASP: An efficient proteomics sample prep method. Read about papers on these topics recently published in 麻豆传媒色情片 & Cellular Proteomics.

Unsheathing the role of myelin lipids in Alzheimer鈥檚 disease
Xianlin Han, an ASBMB Breakthroughs speaker, discussed his pioneering work on lipidomics and the role of sulfatide lipids in Alzheimer's disease.

Ten interesting quotes from the JBC archives
Older papers include archaic quirks and long-abandoned biological concepts. Some show flashes of ideas that grew into their own fields, and others show that some things never change.

Lipid biomarkers hold clues to stroke recovery
Scientists at the University of Arizona found that a lipid mediator accumulates with the waves of inflammation associated with stroke and foamy macrophages.

From the JBC archives: Madness, indoles and mercury-based cathartics
A 1907 paper sought to resolve an ongoing question of whether indole, a bacterial molecule in the gut, could cause insanity if overproduced.

From the journals: JBC
Linking modified cysteines to cell migration. Recognizing protein tags for degradation. Disrupting C. difficile toxin production. Read about recent JBC papers on these topics.

